Generate spatial layers for modelling
Claude Spencer & Brooke Gibbons
2023-11-13
spatial-layers.RmdR set up
Load the necessary libraries.
library(sf)
library(terra)
library(stars)
library(starsExtra)
library(dplyr)
library(tidyr)
library(stringr)
library(here)Set the study name.
name <- 'example-bruv-workflow'Load bathymetry data and create bathymetry derivatives
The bathymetry data used in the example is a relatively low spatial resolution dataset downloaded from https://ecat.ga.gov.au/geonetwork/srv/eng/catalog.search#/metadata/148758. Feel free to replace with any suitable bathymetry in .tif format.
bathy <- rast(here::here("r-workflows/data/spatial/rasters/swc_ga250m.tif")) %>%
clamp(upper = 0, lower = -250, values = F) %>%
trim()Calculate terrain derivatives.
preds <- terrain(bathy, neighbors = 8,
v = c("slope", "aspect", "TPI", "TRI", "roughness"), # Remove here as necessary
unit = "degrees")Calculate detrended bathymetry.
zstar <- st_as_stars(bathy)
detre <- detrend(zstar, parallel = 8)
detre <- as(object = detre, Class = "SpatRaster")
names(detre) <- c("detrended", "lineartrend")Join bathymetry with terrain and detrended derivatives.
Save spatial-layers.
Load metadata and extract site-specific derivatives
Read in the metadata.
metadata <- readRDS(here::here(paste0("r-workflows/data/tidy/",
name, "_metadata.rds"))) %>%
dplyr::mutate(longitude_dd = as.numeric(longitude_dd),
latitude_dd = as.numeric(latitude_dd)) %>%
glimpse()## Rows: 94
## Columns: 12
## $ campaignid <chr> "2022-05_PtCloates_stereo-BRUVS", "2022-05…
## $ sample <chr> "1", "2", "3", "4", "5", "6", "7", "8", "9…
## $ longitude_dd <dbl> 113.5447, 113.5628, 113.5515, 113.5555, 11…
## $ latitude_dd <dbl> -22.7221, -22.6957, -22.7379, -22.7337, -2…
## $ date_time <chr> "2022-05-22T10:03:24+08:00", "2022-05-22T1…
## $ location <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
## $ site <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
## $ depth_m <chr> "93.9", "77.3", "78.3", "73.9", "81.9", "7…
## $ successful_count <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "Yes", …
## $ successful_length <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "Yes", …
## $ successful_habitat_forward <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
## $ successful_habitat_backward <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…Convert the metadata to a spatial file.
Check to see if the metadata and bathymetry align correctly.
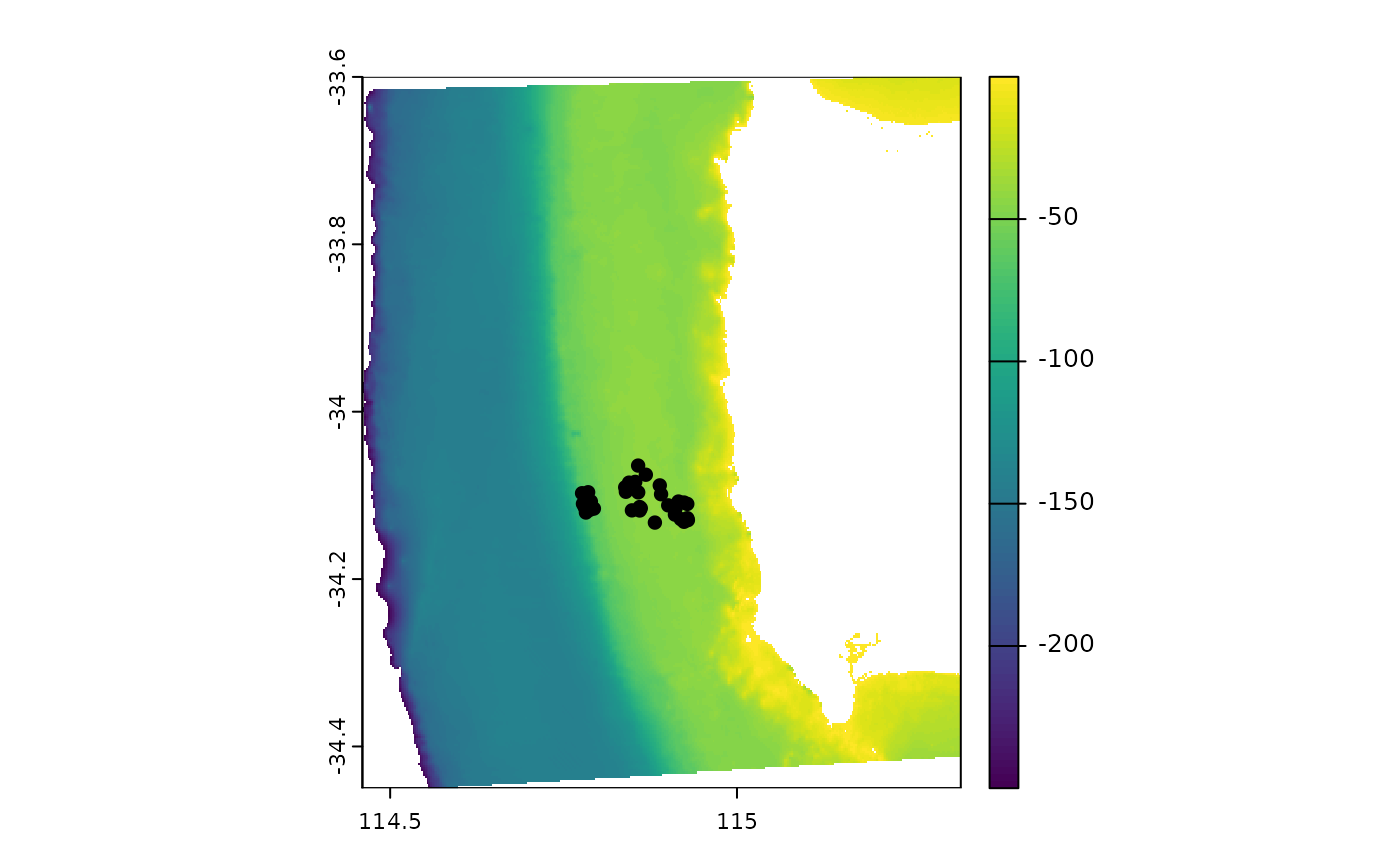 Add the latitude and longitude coordinates back into the metadata.
Add the latitude and longitude coordinates back into the metadata.
tidy.metadata_t <- as.data.frame(metadata.vect, geom = "XY") %>%
left_join(metadata)## Joining with `by = join_by(campaignid, sample, date_time, location, site,
## depth_m, successful_count, successful_length, successful_habitat_forward,
## successful_habitat_backward)`Extract the bathymetry derivatives at each sampling location.
metadata.bathy.derivatives <- cbind(tidy.metadata_t,
terra::extract(preds, metadata.vect)) %>%
dplyr::filter(!is.na(depth),
!is.na(roughness)) %>%
glimpse()## Warning: There was 1 warning in `dplyr::filter()`.
## ℹ In argument: `!is.na(depth)`.
## Caused by warning in `is.na()`:
## ! is.na() applied to non-(list or vector) of type 'closure'## Rows: 32
## Columns: 22
## $ campaignid <chr> "2023-03_SwC_stereo-BRUVs", "2023-03_SwC_s…
## $ sample <chr> "35", "5", "26", "23", "29", "4", "32", "3…
## $ date_time <chr> "14/03/2023 23:36", "14/03/2023 23:49", "1…
## $ location <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
## $ site <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…
## $ depth_m <chr> "39.6", "42.7", "36", "41", "42.6", "45", …
## $ successful_count <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "Yes", …
## $ successful_length <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "Yes", …
## $ successful_habitat_forward <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "Yes", …
## $ successful_habitat_backward <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "Yes", …
## $ x <dbl> 114.9236, 114.9292, 114.9284, 114.9190, 11…
## $ y <dbl> -34.13155, -34.12953, -34.12729, -34.12832…
## $ longitude_dd <dbl> 114.9236, 114.9292, 114.9284, 114.9190, 11…
## $ latitude_dd <dbl> -34.13155, -34.12953, -34.12729, -34.12832…
## $ ID <dbl> 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73…
## $ mbdepth <dbl> -34.97151, -36.35807, -40.68553, -38.25594…
## $ slope <dbl> 0.146843375, 0.812689749, 0.694289634, 0.4…
## $ aspect <dbl> 209.89577, 62.41434, 40.87387, 294.10675, …
## $ TPI <dbl> 0.42153454, 2.39535522, -0.67607403, 0.476…
## $ TRI <dbl> 0.75557327, 3.29823494, 2.39221191, 1.8367…
## $ roughness <dbl> 2.21119308, 8.36493301, 8.36493301, 5.3012…
## $ detrended <dbl> -5.6631737, -7.0394716, -11.2637815, -8.69…